Shotgun cloning, also known as shotgun sequencing, is a widely used technique in molecular biology and genomics to determine the DNA sequence of an organism or a specific region of its genome. The method involves fragmenting the target DNA into smaller pieces, sequencing these fragments, and then assembling the sequenced fragments to reconstruct the original DNA sequence.
The process of shotgun cloning involves several key steps:
- DNA Fragmentation: The target DNA is randomly fragmented into smaller segments. This fragmentation can be achieved using various methods, such as physical shearing, enzymatic digestion, or sonication. The resulting DNA fragments are typically of varying sizes, ranging from a few hundred base pairs to several thousand base pairs.
- Cloning into Vectors: The fragmented DNA is then ligated into cloning vectors, which are small, circular pieces of DNA capable of self-replication. The vectors used are often bacterial plasmids or phages. Each DNA fragment is ligated into multiple copies of the vector, generating a library of cloned fragments.
- Library Amplification: The vector containing the ligated DNA fragments is introduced into a host organism, usually bacteria. The bacteria multiply and amplify the DNA fragments, resulting in a large number of cloned DNA fragments.
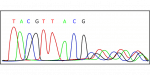
- DNA Sequencing: The next step involves sequencing the cloned DNA fragments. In traditional Sanger sequencing, the fragments are subjected to dideoxynucleotide sequencing using fluorescently labeled nucleotides. More recently, high-throughput sequencing technologies, such as next-generation sequencing (NGS), have been employed, which allow for the simultaneous sequencing of millions of fragments.
- Sequence Assembly: The generated sequences from the sequencer are then computationally analyzed and aligned to identify overlapping regions among the fragments. By identifying the overlapping regions, the sequences can be assembled, and the original DNA sequence can be reconstructed.
- Gap Filling and Validation: In some cases, there may be gaps or ambiguous regions in the assembled sequence. Additional sequencing or targeted PCR amplification may be performed to fill in these gaps and validate the final sequence.
Shotgun cloning has been instrumental in large-scale genome sequencing projects, such as the Human Genome Project. It allows for the rapid and efficient sequencing of complex genomes by breaking them into manageable fragments. Shotgun sequencing has also been applied to the study of microbial genomes, environmental genomics, and metagenomics, among other fields.
Advancements in high-throughput sequencing technologies have made shotgun cloning even more powerful and cost-effective, enabling the sequencing of entire genomes or specific regions of interest in a relatively short time frame. The technique continues to play a crucial role in advancing our understanding of genomics, genetics, and molecular biology.

![DNA. Nucleic acid purification. Okazaki fragments. PCR (digital PCR [dPCR], multiplex digital PCR, qPCR), DNA Repair, DNA Replication in Eukaryotes, DNA replication in prokaryotes, DNA barcoding](https://foodwrite.co.uk/wp-content/uploads/2016/11/dna-geralt-pixaby-03539309_640-150x150.jpg)
Leave a Reply