Proteins are the molecular workhorses of life, and our understanding of their structures and functions has been shaped by the study of a relatively small number of “model” systems. Among these, haemoglobin stands out as uniquely influential. Known primarily as the oxygen-carrying pigment of red blood cells, haemoglobin has been scrutinised by generations of scientists. In the process, it has illuminated fundamental truths about macromolecular architecture, ligand binding, allosteric regulation, conformational change, the genetic basis of disease, and evolutionary adaptation. In this essay, I will explore how the investigation of haemoglobin, from the nineteenth century to the present, has revealed general principles of protein structure and function that extend far beyond the red cell and continue to guide biological thought.
Early Discoveries: From Pigment to Protein
The earliest systematic investigations of haemoglobin can be traced back to Felix Hoppe-Seyler, who in 1862 crystallised the pigment from blood and demonstrated that it was a distinct chemical substance rather than an amorphous colloid. Hoppe-Seyler recognised that haemoglobin was a conjugated protein consisting of a globular polypeptide and an iron-containing haem prosthetic group. Crucially, he demonstrated that haemoglobin bound oxygen reversibly, providing a direct molecular explanation for the blood’s oxygen-carrying capacity. These findings challenged prevailing views of proteins as ill-defined colloids and laid the foundation for the modern concept of proteins as discrete macromolecules with defined composition and function.
By the turn of the twentieth century, physiologists such as Christian Bohr were probing haemoglobin’s oxygen-binding properties. Bohr noted that the oxygen dissociation curve of haemoglobin was sigmoidal rather than hyperbolic, unlike the curve predicted for simple binding according to the law of mass action. This observation implied that oxygen binding at one haem site influenced the affinity of the remaining sites. The concept of cooperativity emerged, and with it the recognition that proteins could act not as passive carriers but as finely tuned molecular machines.
Haemoglobin as a Macromolecule: Establishing Quaternary Structure
In the early twentieth century, a fundamental question was whether proteins were true macromolecules or colloidal aggregates. The answer came from The Svedberg’s pioneering use of analytical ultracentrifugation in the 1930s. He demonstrated that haemoglobin had a molecular mass of ~64,500 daltons, far exceeding that of ordinary metabolites. Moreover, haemoglobin was shown to consist of four subunits, later identified as two α-chains and two β-chains. This was among the first demonstrations of quaternary structure — the principle that many functional proteins are composed of multiple polypeptides arranged into defined higher-order complexes. The notion of proteins as modular assemblies with distinct subunits has since become central to biochemistry, applicable to enzymes, receptors, and structural proteins alike.
Spectroscopy, Cooperativity, and the Bohr Effect
The study of haemoglobin was also instrumental in linking protein structure to physiological regulation. Christian Bohr’s discovery of the so-called Bohr effect — the observation that haemoglobin’s oxygen affinity decreases in response to lowered pH and increased CO₂ concentration — provided one of the first demonstrations of allosteric regulation. Haemoglobin was not merely an oxygen sponge; it was a molecule exquisitely sensitive to its chemical environment. In tissues producing CO₂ and acid, haemoglobin’s affinity for oxygen decreased, facilitating oxygen release where it was most needed. In the lungs, where pH was higher and CO₂ lower, affinity increased, promoting oxygen uptake.
This principle of allostery, first described in haemoglobin, has since been recognised as universal. Enzymes such as aspartate transcarbamoylase, phosphofructokinase, and countless receptors and channels exhibit analogous behaviour, whereby ligands or environmental cues at one site modulate activity at another. Haemoglobin thus provided the first rigorous case study of allosteric control, a concept that has become a cornerstone of protein biochemistry.
Structural Biology: The First Protein Structures
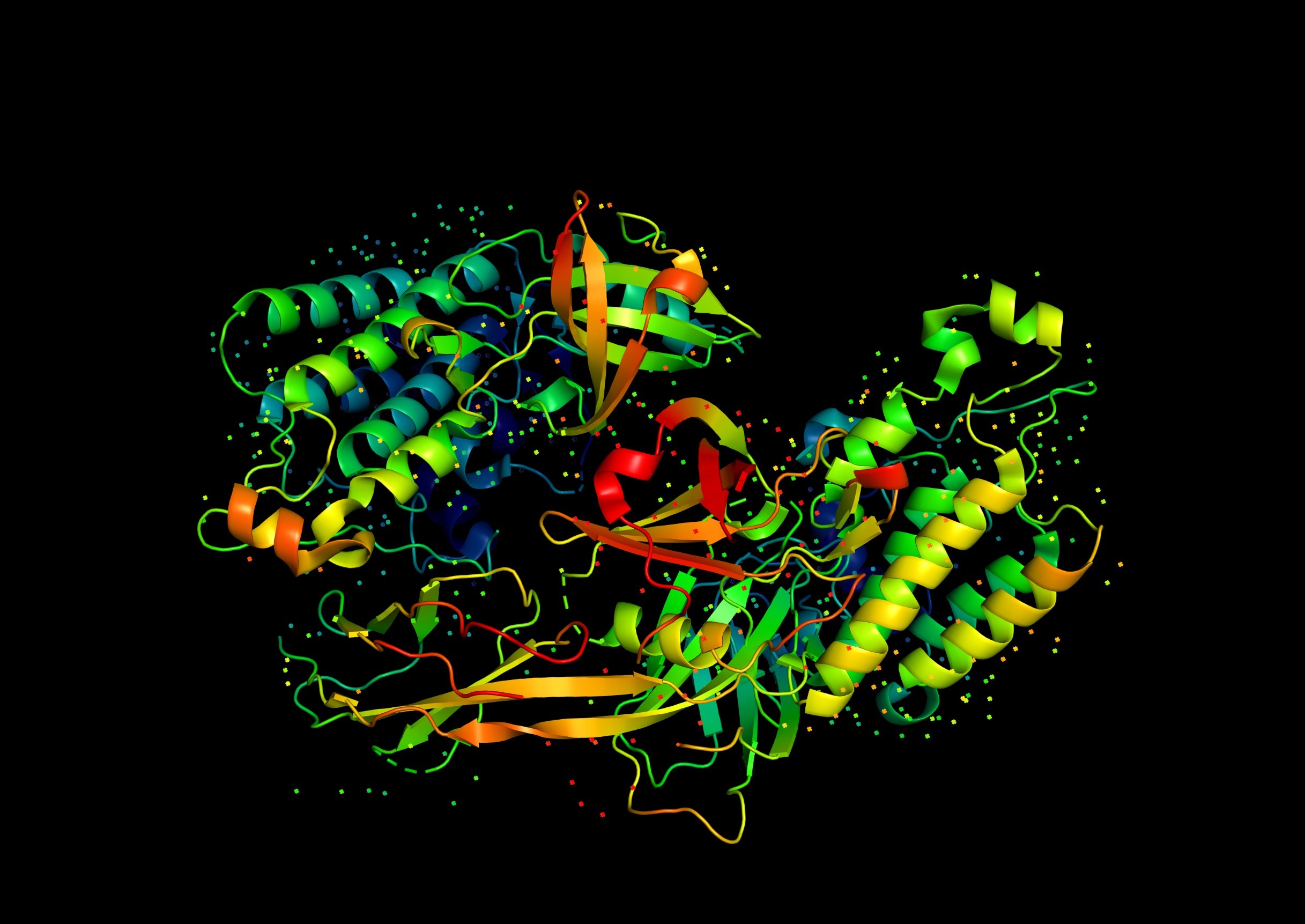
Perhaps the most dramatic contribution of haemoglobin studies came with the advent of structural biology. In the 1950s, Max Perutz and John Kendrew, working at Cambridge, succeeded in determining the first three-dimensional protein structures by X-ray crystallography. Kendrew solved myoglobin, a monomeric oxygen-binding protein, while Perutz revealed the tetrameric structure of haemoglobin in 1959. These structures validated the predictions of Linus Pauling and Robert Corey regarding the α-helix as a common secondary structure element. They also revealed, for the first time, how multiple subunits pack together in a quaternary arrangement.
Perutz’s haemoglobin structures showed that the binding of oxygen induces subtle but concerted conformational changes in the quaternary structure, shifting the molecule between a low-affinity tense (T) state and a high-affinity relaxed (R) state. This provided a structural basis for the earlier physiological observations of cooperativity and the Bohr effect. At a general level, the haemoglobin work established the principle that proteins undergo conformational changes upon ligand binding, and that such changes can be transmitted across subunits to regulate function. This idea has become central to our understanding of enzymes, receptors, ion channels, and signalling proteins.
Models of Allostery: From Haemoglobin to General Theory
Haemoglobin studies not only revealed structural mechanisms of cooperativity but also inspired theoretical frameworks that apply widely across biology. In 1965, Jacques Monod, Jeffries Wyman, and Jean-Pierre Changeux proposed the concerted model (MWC model) of allostery, taking haemoglobin as their prototype. In this model, proteins exist in an equilibrium between two conformational states (T and R), and ligand binding shifts the equilibrium. A year later, Daniel Koshland and colleagues offered the sequential model, in which ligand binding induces stepwise conformational changes propagated across subunits. Although originally debated as competing explanations, these models are now seen as complementary, each applicable in different contexts. Crucially, both originated in the study of haemoglobin, making it the exemplar of cooperative transitions in proteins.
Haem–Protein Interactions: The Role of the Protein Matrix
Another general lesson from haemoglobin is the importance of the protein environment in tuning cofactor chemistry. The haem group can bind oxygen reversibly only when embedded within the globin fold. In free haem, oxygen binding leads to irreversible oxidation of iron to the ferric state, rendering it inactive. The globin fold shields the haem, positions critical residues such as the proximal and distal histidines, and stabilises bound oxygen. This illustrates the principle that cofactors such as haem, flavins, and metal clusters acquire their functional properties only in the context of a specific protein scaffold. The concept of an “active site” shaped by both prosthetic group and surrounding residues is now fundamental to enzymology.
Genetics, Mutations, and the First Molecular Disease
The investigation of haemoglobin was equally pivotal in linking protein structure to genetic information and disease. In 1949, Linus Pauling described sickle cell anaemia as the first molecular disease. Using electrophoresis, he demonstrated that sickle haemoglobin (HbS) had an altered mobility compared to normal haemoglobin (HbA), suggesting a change in charge. A few years later, Vernon Ingram pinpointed the cause: a single amino acid substitution, glutamate to valine at position 6 of the β-chain. This was the first time a single genetic mutation was traced to an altered protein structure and linked to clinical pathology.
This breakthrough established the paradigm that DNA mutations give rise to protein variants with altered structure and function, which in turn can cause disease. Today, this logic underpins the entire field of molecular medicine, from cystic fibrosis to oncogenic mutations. Haemoglobin thus provided the prototype for understanding the molecular basis of genetic disorders.
Evolutionary Biology and Molecular Adaptation
Comparative studies of haemoglobin sequences and structures across species further extended its significance. In the mid-twentieth century, haemoglobin was among the first proteins sequenced in multiple organisms, allowing direct comparisons of amino acid sequences across evolutionary lineages. These studies revealed conserved residues critical for function as well as variable positions associated with adaptation.
Striking examples include the haemoglobins of high-altitude species such as llamas or bar-headed geese, which have higher oxygen affinity to cope with hypoxia. In contrast, diving mammals such as whales and seals possess myoglobins with enhanced solubility to store oxygen during long submersions. Even within humans, fetal haemoglobin (α₂γ₂) has higher oxygen affinity than adult haemoglobin, ensuring effective transfer of oxygen across the placenta. These observations demonstrated how small sequence changes can fine-tune protein function, establishing haemoglobin as a model system for molecular evolution and adaptation.
Protein Dynamics: Haemoglobin as a Prototype
In recent decades, the study of haemoglobin has highlighted another general principle: proteins are dynamic ensembles rather than static structures. Techniques such as nuclear magnetic resonance, time-resolved crystallography, and cryo-electron microscopy have shown that haemoglobin samples multiple conformations even in the absence of ligand. Oxygen binding shifts the population distribution between these states rather than inducing a single rigid transformation. This view, sometimes called the “ensemble model,” has broad implications for understanding enzymes, signalling proteins, and receptors, all of which function through dynamic fluctuations. Once again, haemoglobin provided the first and most thoroughly investigated example.
Biomedical and Biotechnological Applications
The practical implications of haemoglobin studies have also been profound. The measurement of glycated haemoglobin (HbA1c) is now central in the diagnosis and monitoring of diabetes. Attempts to develop haemoglobin-based oxygen carriers (HBOCs) as blood substitutes rely directly on the structural and functional knowledge of the molecule. Protein engineering studies have used haemoglobin as a scaffold to test theories of mutagenesis, stability, and directed evolution. These applications demonstrate how a model protein can serve both as a theoretical paradigm and as a springboard for translational innovation.
General Principles Revealed
Taken together, haemoglobin research has yielded a series of general principles that have become central to protein science:
-
Proteins are discrete macromolecules with defined molecular weights and structures.
-
Quaternary structure enables functional regulation in multisubunit assemblies.
-
Cooperativity and allosteric regulation are universal mechanisms controlling protein function.
-
Cofactor chemistry is tuned by the surrounding protein scaffold.
-
Single mutations can cause dramatic functional changes, explaining molecular disease.
-
Protein evolution involves subtle sequence changes that fine-tune function to ecological niches.
-
Proteins are dynamic ensembles, and function arises from conformational equilibria.
-
Model systems can illuminate general principles across biology, making haemoglobin not only physiologically vital but also conceptually transformative.
Conclusion
From its first crystallisation in the nineteenth century to its role in cutting-edge structural and biophysical research today, haemoglobin has been a touchstone for protein science. It demonstrated that proteins are macromolecules with defined structures, that many function as multisubunit complexes, and that ligand binding can be cooperative and allosterically regulated. It linked single mutations to disease, provided a paradigm for molecular evolution, and showed how protein dynamics underpin biological activity. Few molecules have been studied with such intensity, and few have repaid that attention with such profound lessons. Haemoglobin thus stands as both a vital physiological mediator and a Rosetta stone for deciphering the language of protein structure and function.
References
-
Hoppe-Seyler, F. (1862). Ueber das Crystallinische Globulin. Virchows Arch Pathol Anat.
-
Bohr, C., Hasselbalch, K., & Krogh, A. (1904). Über einen in biologischer Beziehung wichtigen Einfluss, den die Kohlensäurespannung des Blutes auf dessen Sauerstoffbindung übt. Skand Arch Physiol.
-
Svedberg, T. (1930s). Ultracentrifugation studies of haemoglobin.
-
Perutz, M. F. (1960). Structure of haemoglobin. Nature.
-
Monod, J., Wyman, J., & Changeux, J. P. (1965). On the nature of allosteric transitions: A plausible model. J Mol Biol.
-
Ingram, V. M. (1957). Gene mutations in human haemoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature.
-
Dickerson, R. E., & Geis, I. (1983). Hemoglobin: Structure, Function, Evolution, and Pathology. Benjamin/Cummings.
-
Stryer, L. (2019). Biochemistry (9th ed.). W. H. Freeman.
An article based on a question from a University of Bristol exam paper of the 1970s.
Leave a Reply